PL(03/17/14): The revised version of manuscript is here. PCOMPBIOL-D-13-01692_ manuscript_03172014.docx, PCOMPBIOL-D-13-01692_ manuscript_03172014.pdf
PL(03/12/14): One possible way to reduced the number of figures is to combine Figure 9-11 into one. I made a copy under PMC-AT PLIN/Figures named Fig9-11-4FVT_MDs_NADp_local.tif.
PL(03/11/14):
The updated draft are in place under PMC-AT PLIN. All the figures are formatted according to the requirement by PCB and placed under PMC-AT PLIN/Figures.
I will continue to check the draft and update.
RC: Is there any prospect to combine the interaction diagrams with the earlier figures (a,b in each figure, b being the ligand interaction diagram)?
PL: In order to combine interaction diagrams to earlier figures and still make each of them readable, we will need to create large figures. And since these figures shows different perspectives of the structural information, combination of both may actually cause the lost of focus.
RC: Please try to combine the two figures on NAM/isoNAM. The Sir2 ligand interaction diagram can be removed. Due to the smaller size of the ligand interaction diagrams for these, it may be possible. I am trying to reduce the number of figures and will advise further shortly after I check the resulting length. (As indicated earlier Fig 8 may be removed and put in supporting info as well.)
PL(03/12/14): I placed a new version of Figure 11 under PMC-AT PLIN/Figures. Please let me know if that is what you expecting. I still need to make some further correction of the image.
RC: Something like this may work. We would need a,b,c listed on the three parts.
Please make captions for both the new combined figures you made.
PL: Raj, you have to decide which combined figure to use, because combined figure 9-11 and combined figure of 11 and 14 have overlap in figure 11. Therefore we can only chose one to use.
RC: Some computational referee responses that should be reviewed have been posted to PMC-AT PLIN. In particular, the first point about a survey of crystal structures may need to be added. Please also verify that the description in Results of SirTm 2H4F is sufficient per reviewer's comment.
PL(03/10/14):
An updated draft of computational results and methods is in PMC-AT PLIN. I will review together with XG tomorrow to finalize the version.
RC (3/11/14): Ligand interaction diagram figures and associated discussion are posted to PMC-AT PLIN. Please check/edit these as well where necessary.
Some draft discussion of residue contacts to NAM/isoNAM has been added but needs to be checked for consistency against the structures.
To reduce the number of figures, is there any scope for showing the same information on contacts in the ligand interaction diagrams in the earlier figures, or to
make the ligand interaction diagrams parts of the earlier figures?
RC (3-10):
Computational results and methods section draft is in PMC-AT PLIN. Please
- Review for accuracy; edit w MS word revision tracking where appropriate
- Insert binding affinities from tables into specified places in text
- Address comments with underline or in square brackets
- Add citations where specified (XG can finalize with endnote); this includes some MM-PB(GB)SA references
- Make high resolution final versions of the figures if necessary.
- Make sure all text formatting (subscripts,superscripts, etc) is correct
- Find the draft bibliography attached. This includes possible computational references. Please eliminate obsolete old references if they are no longer being cited to make space. Please finalize the computational references.
- Please finalize computational methods sections. For example, it appears that the methods for free energy computations on enzyme, ligand and complex are not described. Also, please review the description of entropy calculations.
- Review several long PCB papers and try to provide an estimated word count for the purpose of length checking
Draft bibliography:
draft_bibliography.docx
RC (3-7): I have put a doc called manuscript figure drafts in PMC-AT PLIN. These are some of the figures I am considering using at this time. The captions need to be
revised since I have taken some of the caption from a larger original figure showing the whole protein and combined it with the more recent ones you provided on the zoomed in figures.
Please finalize the captions. Also, if you like you can change the dynamic SIRT3 binary complex figure to white background for consistency (not sure if the color scheme would work; if not,
it is ok as is).
PL(03/07/14): Updated figure draft in PMC-AT PLIN, including updated figures and captions. Let me know if any further modification is needed.
RC (3/6/14):
Can the following be combined into one table?
NAD+ in SIRT3 and Sir2TM in ternary complex:
| SIRT3/Ac-CS2/ NAD+ |
Sir2TM/Ac-p53/NAD+ |
|
| MM-PBSA |
-5.35 |
-23.19 |
| MM-GBSA |
-65.63 |
-105.48 |
NAD+ in SIRT3 and Sir2TM in complex with NAM:
| SIRT3/Ac-CS2/NAD+/NAM |
Sir2TM/Ac-p53/NAD+/NAM |
|
| MM-PBSA |
-8.10 |
-16.15 |
| MM-GBSA |
-63.12 |
-81.78 |
PL (03/07/14): Table attached. Binding affinities of NAD.docx
RC (3/4/14): Feedback regarding figures for paper:
SIRT3 figures:
Figures showing the whole protein will be very difficult to view when reduced in size. They should be put on white background as in Fig 8a. The latter figure is easier to view.
In addition,
Figure 2a: Observation of A, B and C pockets from the MD averaged structure.
Please prepare a version with NAD in AC pocket superimposed
Figure 3a: Observation of A, B and C pockets from the MD averaged structure.
Please prepare a version with NAM structure superimposed
Sir2 figures:
Use revised versions of Fig 11 or 12 in Sir2Tm fig doc per the types of changes indicated above - whole protein should be put on white background in the style of Fig 8a SIRT3. Also please prepare closeups with contacts and A,B,C pockets shown in the style of Fig 1a/2a/3a, SIRT3.
In general, if a binding mode shows some important contacts that were not observed in a crystal structure used to prepare the complex, please specify residue names and numbers making these contacts. If there is a ligand interaction diagram corresponding to it, this is not absolutely needed; but, we may not be able to include all ligand interaction diagrams in the paper due to space limitations.
PL(03/05/14): Figures updated. The pdf versions are posted here. The original word versions were placed under PMC-AT PLIN. SIRT3_Struct_Align_Figures_MD_vs_Cryst_v1.pdfSir2TM_Struct_Align_Figures_MD_vs_Cryst_v1.pdf
RC: We will need the original versions of all figs in the dropbox to be made into hi-res images (right now some of the writing is not part of the image) for the final draft.
PL: Do you mean you want to merge the circles and notations of A, B and C pocket into the images?
RC: Yes. I believe there are also some resolution requirements for the journal, you may check with XG.
PL(03/06/14): An updated version of the two files are now under PMC-AT PLIN. I will check with XG about the resolution issue.
PL(03/06/14): One table showing the changes in residues in contact with NAD+ in various complexes. The current cutoff distance is set to 3.5 Angstrom, while previous cutoff distance in preparation of interactions diagram is 4 Angstrom. Therefore, all the in-contact residues should be visible in the interaction diagram. I am working on the residue-NAD+ MM interaction energy analysis which should provide more accurate assessment of the important contacts. (The interaction diagrams included in this file is for visualization of contacts only, and there are not for actual use.) Again, only pdf format is attached and the original word document is under PMC-AT PLIN in Dropbox. Residues_in_contacts.pdf
PL (03/03/14): List of systems modeled and the short description about the preparation steps. System modeled.docx
RC (2/23/14): Comments regarding schedule/tasks (breakdown of tasks under 1 above, which is priority):
a) Figures: NAM/isoNAM ligand interaction diagrams needed.
Edits to other figures may be requested shortly.
b) Manuscript discussion: discussion will consider relative binding affinities of NAD+ and NAM, in SIRT3 and Sir2 AC pockets of ternary complexes. Provide bullet points comparing these binding affinities and the relevant interactions visible in the average MD figures as they relate to the MM-PB(GB)SA energy breakdown by interaction type. This will be used to replace the discussion written by Eric on the comparison of protein:NAD+interactions in SIRT3 and Sir2.
c) Manuscript discussion: related to b), for each of the complexes for which binding affinities are reported and which were discussed in the original paper draft, provide bullet points regarding the relevant interactions visible in the figures as they relate to the MM-PB(GB)SA energy breakdown by interaction type.
d) Manuscript discussion: Provide bullet points comparing the fluctuations in the MD trajectories for NAD+ in the SIRT3 and Sir2Tm ternary complexes (relt'd to b). If entropy was calculated for the complexes which equilibrated, this could be used as a point of comparison of the distributions.
e) Manuscript discussion: Provide the requested average MD figure for a snapshot other than the last 10 ps which shows another mode of the distribution, in order to assess how much discussion should be devoted to the interactions in the average MD figures.
f) Methods: Pending methods sections should be completed
g) Methods: Please confirm that entropy is included in the reported binding affinities. MD convergence analysis: would like more details to be provided on how simulation time was set/how convergence of MM-PB(GB)SA calculations was assessed (or a figure with caption to be prepared showing the convergence rate of the MM-PB(GB)SA binding affinities with simulation time for a couple of complexes)
h) Next paper (after all the above are done): Begin to set up SIRT3 C pocket MD binding affinity calculation for the tightest binding small molecule identified by XG (for which charge fitting has already been done)
i) Next paper ("): Comment on the comparison of MD trajectories for NAM in Sir2Tm vs SIRT3, with respect to the role of Met70.
j) Next paper ("): Begin to set up NAM/isoNAM C pocket MD simulations for SIRT1. Further details will be discussed when you are ready.
PL(02/27/14): Component analysis and some discussion related to b-d) above is provided in the attached document.
Let me know if this is something you are looking for.
PL(03/03/14): Updated component analysis with NAD+ and isoNAM binding in SIRT3/Ac-CS2/NAD+/isoNAM system.Binding affinities discussion.docx
RC (02/28/14): Yes, this is what I was asking for. Please provide isoNAM binding affinity data in the same table as NAM,
I am reviewing bullet points for energy component /structural analysis and will advise as task early next week if any additional points regarding residue contacts will be useful in the replacement of the corresponding discussions in Eric’s original discussion sections.
Given the various additions to the draft, including energy component analysis and additional complexes simulated, this may not be necessary.
related to d) and g) No, I did not carry out entropy calculations. The NMODE analysis is too time-consuming and quasi-harmonic approximation is not accurate.
Still working on the rest of the requests.
PL(02/28/14): Analysis regarding requests d-e) is provided in the attached document. MD_trajectory discussion.docx
RC (2/28/14): This analysis is useful; will look over/consider how to incorporate in manuscript and advise whether any tabular presentation of trajectory statistics will be advisable as task for early next week.
PL(02/28/14): Computational method section with updated references. Methods_comp.docx
PL(02/25/14): Update figures for MD structural analysis of SIRT3/Ac-CS2/NAD+, SIRT3/Ac-CS2/NAD+/NAM,SIRT3/Ac-CS2/NAD+/isoNAM, SIRT3/NAD+; interactions diagrams include SIRT3-NAD+, SIRT3-NAM and SIRT3-isoNAM (residue numbers were corrected). SIRT3_Struct_Align_Figures_MD_vs_Cryst_v0.pdf
Update figures for MD structural analysis of Sir2TM/Ac-p53/NAD+, Sir2TM/Ac-p53/NAD+/NAM, Sir2TM/NAD+; interactions diagrams include Sir2TM-NAD+ and Sir2TM-NAM (residue numbers were corrected).Sir2TM_Struct_Align_Figures_MD_vs_Cryst_v0.pdf
(Due to size of the original word files, only PDF files were attached. Original word files are available under Dropbox/PMC-AT Reseach,)
RC: Ok, I have joined that folder now. It may take some time to sync, I will let you know if I am not able to find these files. I was already a member of the original folder, but have joined the new one without leaving the old one. So far I have not seen files with the above names. You may consider also putting them in PMC-AT PLin.
PL (03/04/14): The two files are now also under the directory of PMC-AT PLIN.
RC (02/28/14): Am analyzing structures and will advise what final figures need preparation as task for early next week.
We will not have space to present all these MD results in the current paper, so we will save some for the next paper. Will decide on this shortly.
Although this information is present elsewhere on the wiki, it may be useful to have in one table the pdb entries used as starting structures for each MD simulation, how the ligand complex was prepared (e.g., by alignment with another structure such as Sir2) and (as third column) what structures were compared to in each case for the rmsd analysis).
PL(02/19/14): Figures for structural alignment and binding pocket interactions for Sir2TM system (currently Sir2TM/Ac-p53/NAD+ and Sir2TM/Ac-p53/NAD+/NAM) is attached.
An update version includes binary Sir2TM/NAD+ system is attached.
And binary affinity calculations for Sir2TM is also updated. Binding affinity calculations from MD_Sir2TM.docx
RC (2/23/14): Comments: a) There is a comparison between Sir2Af2/NAD/NAM and Sir2TM/Ac-p53/NAD/NAM reported. For Sir2Af2 (1YC2), does the structure include PEG? Previous docking studies were not able to identify a Sir2 AB pocket binding pose in presence of peptide. By comparison, in SIRT3, docking was able to locate a pose in AX pocket in presence of peptide. (This is relevant to the discussion of this binding mode in Sir2Tm).
b) Binding affinities: did you find that NAD+ binding affinity is more favorable in Sir2TM/NAD+ binary complex than in the ternary complex?
RC: Please provide your comments on the above if they have not been addressed by recent updates. For b, I may have misread the table.
PL(02/28/14): To a) For Sir2Af2 (1YC2), yes, the structure include PEG. Using 4FVT as receptor, we are actually able to find a pose in AB pocket in the presence of peptide (I think it is not necessary to name it as AX pocket, because although it is somewhat different from AB pose from Sir2Af2, we can simply classify this type of binding as AB binding mode). Previous docking studies may not have exhausted all possible conformation search, which could lead to the failure of identifying such binding mode.
To b). Yes. NAD+ binding affinity is more favorable in Sir2TM/NAD+ binary complex than the ternary complex, especially in term of MM-PBSA values. The two values are much closer in MM-GBSA.
PL(02/18/14): Updated information on MM-PB(GB)SA results on SIRT3 and Sir2TM. (Not the final results because MD simulations are not complete for Sir2TM).
Binding affinity calculations from MD_SIRT3.docx
On top of the report are the MM-PB(GB)SA values, and the time-series breakdown in every 2ns and stand deviation and standard error of the mean are followed. The time-series breakdown can be used to keep track on the fluctuations and convergence.
RC (2/9/14): Figures needed for the PCB manuscript:
- 3D representations. Superimpose crystal structure and report RMSD of ligand (MD average to crystallographic) wherever possible. Complete captions indicating method of preparation
n SIRT3:peptide:NAD AX and AC pocket MD averaged structures (parts a,b of figure) and SIRT3:NAD AC pocket (part c of figure)
n SIRT3:NAM/isoNAM: superimposed averaged MD structures in part a of figure. Sir2Tm:NAM averaged MD structure compared to crystallographic structure in part b of figure. Compare binding affinities for both NAM and isoNAM in SIRT3, indicating NAM,isoNAM are congeneric series and binding affinities can be compared
n Sir2:peptide:NAD AC pocket averaged MD structure, comparison to crystal structure.
- Ligand interaction diagrams for all the above (MD average only). Please confirm that all the ligand interaction diagrams prepared by Eric have a corresponding figure based on MD simulations. Binding affinities and energy component analysis will be requested thereafter.
PL(2/14/14): Tasks are not complete so far. The attached are the results I can obtained so far. Figures for Structural alignments:
Computational details:
I will continue to work on it next week.
PL (02/21/14): An updated version of computational method section: Methods_comp.docx
I will continue to update and add reference to it.
RC (2/18/14): a) loop building method in Sir2Tm; b) different approaches to free ligand, protein free energy calculations should be included in the methods.
PL(02/19/14): will do.
PL(02/17/14): An updated version of the structural alignment and binding pocket representation. Struct_Align_Figures_MD_vs_Cryst.docx
The preparation is not in the exact order as request. I presented various representations so you can pick the most suitable combinations.
Analysis of Sir2TM is underway. And ligand interaction diagrams will be prepared at the same time.
Just to clarify, the SIRT3:NAM/isoNAM or Sir2TM:NAM systems include NAD+ and acetylated peptide substrate (Ac-CS2 in SIRT3, Ac-p53 in Sir2TM) adopted from crystal structures.
RC(02/18/14): Are you planning to provide PBSA results for all complexes? At least one figure lists MM-GBSA energies.
PL(02/19/14): On the binding affinity results presented above, I included both MM-PBSA and MM-GBSA values. Some studies suggested that MM-PBSA values may come close to the experimental binding affinity, but MM-GBSA correlates better with experimental values, especially for congenic series. I presented the MM-GBSA values when comparing NAM vs isoNAM. But we can present them all, or using one set and leave the other in the supporting materials.
RC (02/17/14):
a) The NAM/isoNAM/xtal superposition is cluttered. The aligned figures for NAM/isoNAM provided today are preferred. Also in Figs 2a,3a, it appears the perspective was different or the structures were not aligned well, so the orientations of NAM/isoNAM look quite different, but in fact they are found to be very similar upon proper alignment.
You can replace the NAM/isoNAM/xtal superposition figure with just NAM/xtal for clarify. We will decide later whether or not to use this figure.
b) NAD AX / xtal comparison may not be useful since there is no NAD in AX from xtal to compare to. We may replace this with a figure with isoNAM or NAM in C pocket and with NAD in AX/AB to show how all three can bind simultaneously (NAD unproductively), but omitting the xtal structure.
c) Based on your latest posting I understand that the close-ups of the binding pockets are not meant to serve as ligand interaction diagrams, and that you will be providing ligand interaction diagrams in the format previous prepared by Eric. These will be important for generating text descriptions of which residues are involved in interactions.
d) Methods and figure captions indicate use of 4FVT as starting structure for both the ternary and binary complexes of SIRT3. 4FVT pdb structure includes peptide. 3GLS is a structure without peptide. How long did you run this simulation to ensure the domain motions relevant to peptide binding/release were accounted for? This is related to a question asked earlier about the MD simulation for the binary complex, as well as the structural superposition of SIRT3 pockets with/without peptide.
e) The MD structural averages are based on the last 10 ps. However the binding affinities previously reported are based on averages over 10 ns. What was the basis for the choice of 10 ps for the structural averages?
f) Once the choice is finalized, the figures can be arranged according the requested subfigures with captions for a,b,c etc.
PL(02/17/14): More figures were added to the files above. (file updated with same name.)
(a) In new Figure 7, close-up views for binding pockets are presented for various complex at the same perspective, so that you can get a better idea what were changes. (The previous figures were prepared individually and rotated to get a better view in each case.)
Figures comparing NAM/isoNAM/xtal were prepared before in Figure 2 and 3.
(b) A comparison between AX/AB was made by comparing the superimposed AB pose (as in starting structure for complexes with NAM), as presented in Figure 8 and 8a. Let me know if it works for you.
(c) Interaction diagrams were presented in Figure 9.
(d) The MD simulations for binary structure (SIRT3/NAD+) is tricky. I have two settings, but currently only one was presented. The first one start from the 4FVT xtal structure by removing peptide substrate. The second one starts with SIRT3 from 4FVT xtal and NAD+ from 1YC2:A in AB pose. Only the first one was reported. The NAD+ varies dramatically over the course of MD. The presented figure can be viewed as a snapshot of the trajectory instead of a full representation of the dynamic properties. The domain motions are usually much slower than the nano-second range that we are simulating. I didn't try to ensure all the slow motions were sampled. However, since we always started from the ternary xtal structure (in the peptide bounded form.), most of the interactions needed for peptide binding are in place.
RC: As I recall the binary structure was used to estimate the binding affinity in absence of peptide. If the domain motions are much slower than the nanosecond range, and the starting structure was taken from a peptide bound complex, wouldn't this imply that the domain conformations used to estimate the binding affinity were representative of the peptide bound complex?
PL:(02/18/14): As you can see from the SIRT3_NADp_MD_snapshots.docx file I posted on 01/30/14, although we started from the peptide bound complex, and NAD+'s initial structure from AC or AB pose, as the MD continues, the structure evolves too. The changes may not be all completed in term of domain movement, however, some interactions that used to hold peptide may no longer exist, and lead to structural change in the peptide binding pocket, which will will certainly have significant impact on NAD+ binding. If you check out the snapshots for the binary system and final snapshot as presented in the prepared figure, you will see NAD+ is completely out of C pocket. If you check out the MM-PB(GB)SA results, you will see large variations over the course of the MD. Although there is no experimental structural information available for binary complex, we can still draw some reasonable conclusion from MD simulation. 1) NAD+'s adenine binds well to the A pocket, the nicotinamide moiety do not. 2) NAD+ binding in SIRT3 can not be presented by a single static structure and the binding affinity can not be accurately estimated using a short simulation.
RC; Regarding experimental structural information, why didn't you consider use of 3GLS? When building a binary complex with either apo or ternary complex options for starting structures, knowing that the peptide binding induces a significant domain shift, an argument needs to be presented for why the ternary complex was chosen as the starting structure. (As I recall NAD+ binding results primarily in loop conformational changes, which may also be difficult to model.) Please comment.
PL(02/18/14): In the energy landscape, trying to model a bound structure from apo-enzyme usually is a much longer process than to model a unbound structure from a bound one (in this case, we can consider ternary to binary is a process releasing peptide substrate.) The NAD+ binding is not simply about the loop conformational change. Peptide substrate is probably the main driving force to bring the
Rossmann fold domain and Zn-binding domain together. Without that, the C pocket will not properly form. When substrate binds, some loop becomes static, some many still be flexible. In our models, the dynamic loop conformation is system specific. Further structural and energetic analysis is needed to be more specific about its role in NAD+ binding.
RC: The analysis depends on the appropriate interpretation of the simulation. It is not appropriate to consider this an equilibrium simulation of a binary structure. NAD+ binding itself does not result in the domains coming together. The loop adopts a more rigid structure in some sirtuins after NAD binding. We know that peptide binding is the driving force. That is why the issue of the choice of starting structure without peptide is important to clarify. This is why questions were raised earlier about the statistics from this simulation. I will consider the appropriate presentation of the results and ask for more information on the simulation and structures when needed.
(e) The system were in dynamic motion throughout the MD process. Averaging over a long period of time will only produce washout and/or wield structures, e.g. hydrogens of methyl group become one as they can almost rotate freely, and flexible regions will be averaged to a non-nature feature, e.g. a sine wave become a flat line after averaging. The short average over 10ps presents a snapshot of the MD process, which can still be used to understand the interactions between ligands and proteins, although multiple snapshots may be needed in complicated cases.
RC: Ok, so a major reason for using a short snapshot is that unphysical structures could be produced otherwise. For example, you might obtain a structure that is energetically highly unfavorable if the distribution of states is multimodal - since the average could lie between two lower energy local optima. I assume you've looked at longer snapshots and confirmed that this happens in many cases for our structures.
PL (02/18/24): Yes, you can easily find one example from SIRT3_NADp_MD_snapshots.docx posted on 01/30/14.
RC: Please provide an example of such a structure average figure from one of the other modes of the distribution. I would like to see the differences before committing to a structural analysis in the paper based on a particular snapshot.
RC (2/14): XG, I need copies of the relative inhibition figures in an image format that can be used in the manuscript (not pdf). Also, you can include a complete caption with these figures that I will edit. I will let you know shortly what other experimental figures/figure edits I need.
PL(02/07/14): Component analysis for MM-PB(GB)SA results for SIRT3/Peptide/NAD+ system.
(Analysis for other systems will be carried out and updated once available.)
Please pay special attention to the values of last line, which are MM-PB(GB)SA values for free SIRT2, peptide, and NAD+ (MD carried out on free systems). The complex (_com) in the table includes all three components and receptor includes two components (SIRT3 and the other ligand), and the overall energies for both continue to get lower as MD runs for longer.
While the peptides in the complex has a MM-PB(GB)SA valule similar to free peptide, the NAD+ MM-PB(GB)SAis about 20 kcal/mol higher, probably due to the strain in the formation of AC pose.
When NAD+ in the AC pose, there is always an water bridging the interaction between nicotinamide moiety and SIRT3 backbone, which can contribute about 2 kcal/mol in its binding affinity, which is not included in our calculations.
Binding affinity Component Analysis_1.docx
RC (02/18/14): Regarding the water molecule in the AC pocket, I believe you mean there was a bridging crystal water to carba-NAD in the 4FVT structure that was removed in the structure preparation step for MD simulations. Was it also observed in any other available sirtuin structures with cocrystallized NAD or analogues, and was it present in Sir2 structure with free NAM as well?
PL(02/19/14): Yes. I have seem it in ternary structures of SIRT3 (4FVT) and Sir2TM (2H4F) and in 1YC2 chain B and C where NAD+ in AC pocket. However, the O and N of nicotinamide group exchange positions in 1YC2 (could be a mistake). The bridging water also appears in hSIRT5 ternary structure (3RIY and 4G1C). All the waters were removed in structure preparation step for MD simulation. In SIRT3 MD simulations, I have found the water always fills the same spot after a short MD run. For 1YC2 chain A and D (Sir2Af2 with NAM in the C pocket), the bridging water has not been identified.
PL(02/05/14): Some structural analysis from MD simulation: comparison with crystal structures.
Structural Analysis of NAD+ Binding in Sirtuins.pdf
PL (01/28/14) Current status on MM-PB(GB)SA results from MD simulations.
(02/04/14) Updated MM-PB(GB)SA results from MD simulations.
Binding affinity calculations from MD.docx
RC (1-29): Are you running all MD simulations for 25 ns? Or, is convergence analysis used to determine the simulation time? The attached reports the Delta G estimates computed using data over successive intervals of time, but does not describe the convergence of the Delta G's computed using all the data up to a particular time. As with any method for sampling from a distribution, one expects to see fluctuations that increase as the sampling time decreases. How are you planning to use the data on in the attached to assess convergence, and how will you describe this methodology?
Regarding the task below on reporting the breakdown of MM-PB(GB)SA energies into component terms, are you recording that data for each frame when you are carrying out the MM-PB(GB)SA calculations reported herein, so it can be used later?
PL(01/29/14): 1) No. The MD simulations run for various lengths, from 10 ns to 26 ns as of now. 2) Yes. Delta G's were calculated after 1 to 2 ns of preliminary equilibration. MD simulations were extended when significant changes in Delta G (every 2 ns) were found. The time required to achieve equilibrium depends significantly on the system and the starting structure, and it is hard to determine when equilibration is reached, I have used the last 10 ns of the simulation and reported in the mean value. The equilibration and convergence process is most obvious in the Delta G calculations for NAD+, NAM and isoNAM.
Depends on what information we want to deliver in the manuscript, we might report only the mean value for the last 10 ns (or 8 ns) as the calculated Delta G's. And I probably won't report the details about convergence, and simply state that we equlibrate the systems after various ns and Delta G's were obtained for 10 ns (or 8 ns) production runs.
The components of MM-PB(GB)SA energies (per frame) are available, and there are a lot of data. If they are useful to our manuscript, we can extract those out and re-analyze when necessary.
Here are some references about the performance of MM-PB-(GB)SA calculations. ci100275a.pdf21666_ftp.pdfjp404160y.pdf
RC (1-27): Please provide your weekly schedule requested below before EOD (working day) today (Mon 1-27).
See also new tasks below.
RC (1-14): Plan updates based on meeting.
Experimental -
-- Based on information regarding ITC operating range, it is important that we determine whether ITC can measure NAD binding affinity to SIRT1 in absence of peptide as soon as possible, given that data suggests that NAD-SIRT1 is the tightest binding complex (in presence of peptide) that we are currently looking at. Since the experiment may not work, we need to leave time for other experiments (below).
-- Since peptide binding affinity to SIRT3 has already been determined by ITC, we will not consider it at this time
-- If the above is not possible, we will move on to inhibition kinetic studies with isoNAM/SIRT3. Please let me know whether it would take a month for just SIRT3 or whether you meant 1 month for both SIRT1 and SIRT3.
-- Prior to doing inhibition kinetics studies, we should finish the relative inhibition experiments with isoNAM.
-- Please provide a schedule for the above studies so I can determine whether they will fit within our allotted time. If not, we may rearrange tasks.
XG(1-14):
* Depending on how ITC works, measurement of binding constant of NAD-SIRT1 will take 2-3 days.
RC (1-15): As soon as we have the NAD-SIRT1 results, we may need to move directly to inhibition kinetics experiments. Please let me know the estimated date of completion of this experiment once we have the results from the test kit experiment.
XG (1-16): EDTA test does not perform as what it's indicated from provided data sheet (come with the kit). The data along with questions will be sent to GE technical support for future assist. It needs possiblly a mechanical calibration for VP-ITC.
* The relative inhibition experiments with isoNAM: I have finished the determination of solubility of isoNAM, the preparation of isoNAM stock solution, and redone the standard curve. The rest of the experiment will take 3 days.
RC (1-15): Ok, I assume this work was also necessary for the inhibition kinetics experiments below. Please let me know before you start these experiments, so we can decide based on the time we have whether to move directly to inhibition kinetics.
XG (1-16): I prefer to start with the relative inhibition experiments with isoNAM. In this experiment, great excess of isoNAM will be used which will increase the ionic strength of buffer solution. We would like to know if under such condition the Developer II still works fine. The feedback from this experiment will provide a guide of the selection of isoNAM concentration for kinetic study.
* It would take about 3 weeks to finish one enzyme (SIRT1/SIRT3). Then 1 week to analysis the data for both SIRT1 and SIRT3. Total 7 weeks.
RC (1-15): Ok, we don't have time to do both SIRT1/SIRT3 in this way.
How long would it take to run each inhibitor concentration (e.g., high) for one enzyme? Note we already have the [I]=0 data. How many inhibitor concentrations are you assuming above?
XG(1-16): 3 or 4 inhibitor concentrations, will be picked from following concentrations: 500 uM, 1 mM, 10 mM, 25 mM, and 50 mM. Under every inhibitor concentration, five [NAD+] = 100, 375, 750, 1500, 3000uM will be used with fixed [peptide]. 3.5 days for one inhibitor concentration. The above 3 weeks is for testing 4 inhibitor concentrations. It will be nice
actually to have both SIRT1 and SIRT3 tested for the purpose of comparison. Then, if we test 3 inhibitor concentrations for SIRT3 and SIRT1, we would have time to finish both.
RC (1-21): Ok, please remember to update me after 2 concentrations of inhibitor are done.
XG (1-20): Schedule for week of 1/20/14-1/24/14
01.17.2014 – 01.21.2014: relative inhibition study
01.22.2014: Data analysis
01.23.2014: Choice 1-start isoNAM kinetics study (3 concentration, 12 days); Choice 2 - ITC revisit (1 or 2 dsys).
RC (1-21): Ok, I will let you know by EOD Wed whether to continue with isoNAM kinetics study or revisit ITC. Would it be disruptive to pause the isoNAM kinetics experiments next week to revisit ITC?
I was informed that accessories will be delivered in the next few days. It would be best to do these experiments after the accessories arrive. So if pausing the isoNAM kinetics experiments is not an issue, the ITC can be done next week.
RC (1-27): Please provide an updated schedule for this week. Please provide a more detailed schedule including when you will be doing the ITC analysis (this week or next week), additional fitting tasks requested, review of CJ's Prism protocol posted today to wiki, etc.
XG (1-28): 1/27/2014-1/31/2014 isoNAM kinetic study (10 mM and 25 mM). IsoNAM kinetics studies use end-point assay. It includes experiment planning, sample preparation, loading sample, quenching, signal developing, and plate reading. Because it is a very expensive assay, full attention needs to be performed. From last week, my working hour had been extended from 7:00 AM - 6:00PM or even later. By doing so, every isoNAM concentration needs 3.5 days. We are planning to do 3 concentrations, which means the earliest time for ITC evaluation is late next week, say Thursday (Feb 6, 2014). For the requested fitting task, I need to stay extra longer, whenever the day is not packed too full. For CJ's Prism protocol, have downloaded the file, will find time to review.
RC (1-29): Ok, please post the isoNAM data as it becomes available.
RC (2-3): Please plot and post the latest isoNAM data when it becomes available.
Computational -
-- We have already run MD binding affinity calculations on SIRT3/peptide/NAD/NAM. Based on the fact that ITC cannot accurately measure binding affinities for NAM/isoNAM, and the fact that there are no new contacts observed between intermediate and NAM/isoNAM, if XG confirms that it is not possible to directly measure mM binding affinities experimentally with other techniques, there is no particular reason to start binding affinity calculations on the intermediate complexes at this time. Therefore we should dedicate as much GPU time as possible to the SIRT3/peptide/NAD/isoNAM complex to start.
-- Regarding isoNAM as a base exchange substrate, please post the paper by Denu on base exchange substrates or point out where it is on the wiki.
-- Given that it is likely not possible to determine NAD binding affinity to SIRT3 in absence of peptide with ITC, I would like to consider whether it will be possible to run an MD calculation on NAD in AC pocket in absence of peptide, for comparison to the ternary complex already studied. The aim of this study would be to provide some explanation from a computational standpoint as to why the binding affinity changes substantially in absence of peptide. Please let me know if we have the required apo crystal structure and if we can prepare an appropriate starting structure for this calculation.
-- If the latter is not possible, the next MD calculations to do after the SIRT3/peptide/NAD/isoNAM complex should be NAM/isoNAM with 1YC2.
-- Similarly, please provide schedule of work so we can determine how the computational schedule will align with that for experiments.
-- Protocols for the MD binding affinity calculations should be written in a format that can be used in the paper.
RC (1-24): New alignment tasks for computation: a) structure alignment of mouse and human SIRT1; b) structure alignment of mouse and human SIRT3. Important because mouse enzymes have been studied in the prior literature. XG has done sequence alignment of mouse and human SIRT1, and should do the same for mouse and human SIRT3. Results of all studies should be posted here.
XG(1-24): Sequence alignments.
1. Sir2alpha [Mus musculus] vs. Sir2alpha [homo sapiens]
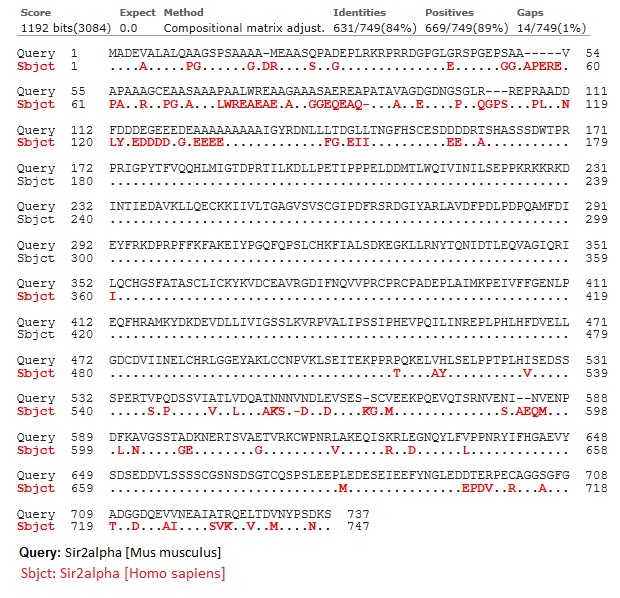
Note:
2. SIRT3[Mus musculus] vs. SIRT3 [homo sapiens]
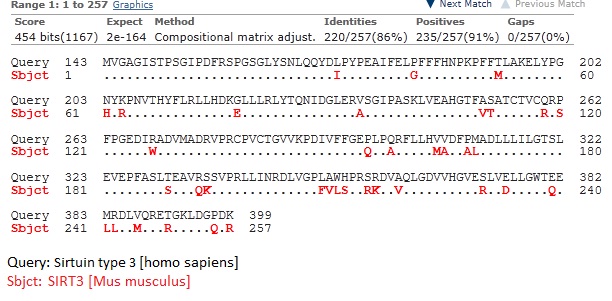
Note: Human SIRT3 (hSIT3) has been reported to be a 399 amino acid protein with two length. The full length hSIRT3 is enzymatically inactive and majorly locates at mitochondria. The smaller 28kDa (257 aa) protein were present in the cytoplasm and nucleus, it becomes activated.
Mouse SIRT3 is a 257 aa protein that aligned well with C-terminal protion of hSORT3 (residues 143-399) and localized in certain areas of the cytoplasm. (Jin L, etal. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Science 18 (2009) 514-525.)
PL (01/24/14): There is no crystal structure available for mouse SIRT1 or SIRT3. And only a limited number of crystal structures available for human SIRT1 as well. With the high sequence similarity between mouse and human sirtuins, we can expect them to have high structure similarity.
RC (1/24): Based on the sequence alignments and the structures of hSIRT1 and hSIRT3, it should be possible to quantify sequence similarity in the pockets of interest (A,B,C, peptide binding pockets) for mouse/human proteins, strengthening this argument for the purposes of our mechanistic study. Please do so. This is related to the task on structural similarity of SIRT1 and Sir2 A,B,C pockets above.
PL (02/03/14): Here is some detailed information about the binding pockets, A, B and C, from structural information for ternary structure in Sir2Af2, Sir2TM, SIRT3, SIRT1. The A and C binding pocket are reasonably similar among all these structures. B pocket can not be clearly defined, as nicotinamide moiety is mostly exposed to the solvent in this position. The catalytic domain of SIRT1 (241-512 as in human) between mouse and human are almost identical except Ile360. And catalytic domain of SIRT3 (122-395 as in human) between mouse and human has lower sequence identify as seen from above.
Alignment_and NAD_binding2.pdf
RC (2/3/14): Does this mean that it is not appropriate to refer to an AB pocket in Sir2 and SIRT1 but an AX pocket in SIRT3?
PL(02/04/14): I think we can still refer to B as a binding pocket, but instead of an usual binding site that defined by a set of residues in contact, this B pocket is open and exposed to solvents. And we should also note that the position of B (or nicotinamide) is not precisely defined as well (not necessary the same position as in 1YC2 chain A or D), and it depends on the size of the side chains at +1 and +2 residues of substrate peptide.
RC (1/27): a) Please provide the converged values of the component terms of the MM-PB(GB)SA energies for the completed binding affinity simulations below.
b) Please provide sequence/structure alignments of SIRT1/Sir2Tm and SIRT3/Sir2Tm.
c) Please verify that Sir2Af2 has Met70 in C pocket. I will indicate later why I am interested in this.
d) Please provide an updated schedule for this week.
PL(01/30/14):
a) Here is an example of the component terms for the MM-GBSA calculations of a 2 ns simulations for SIRT3/NAD+/AcCS2 ternary system.
The notation should be pretty easy to understand. Let me know if you need more explanation.
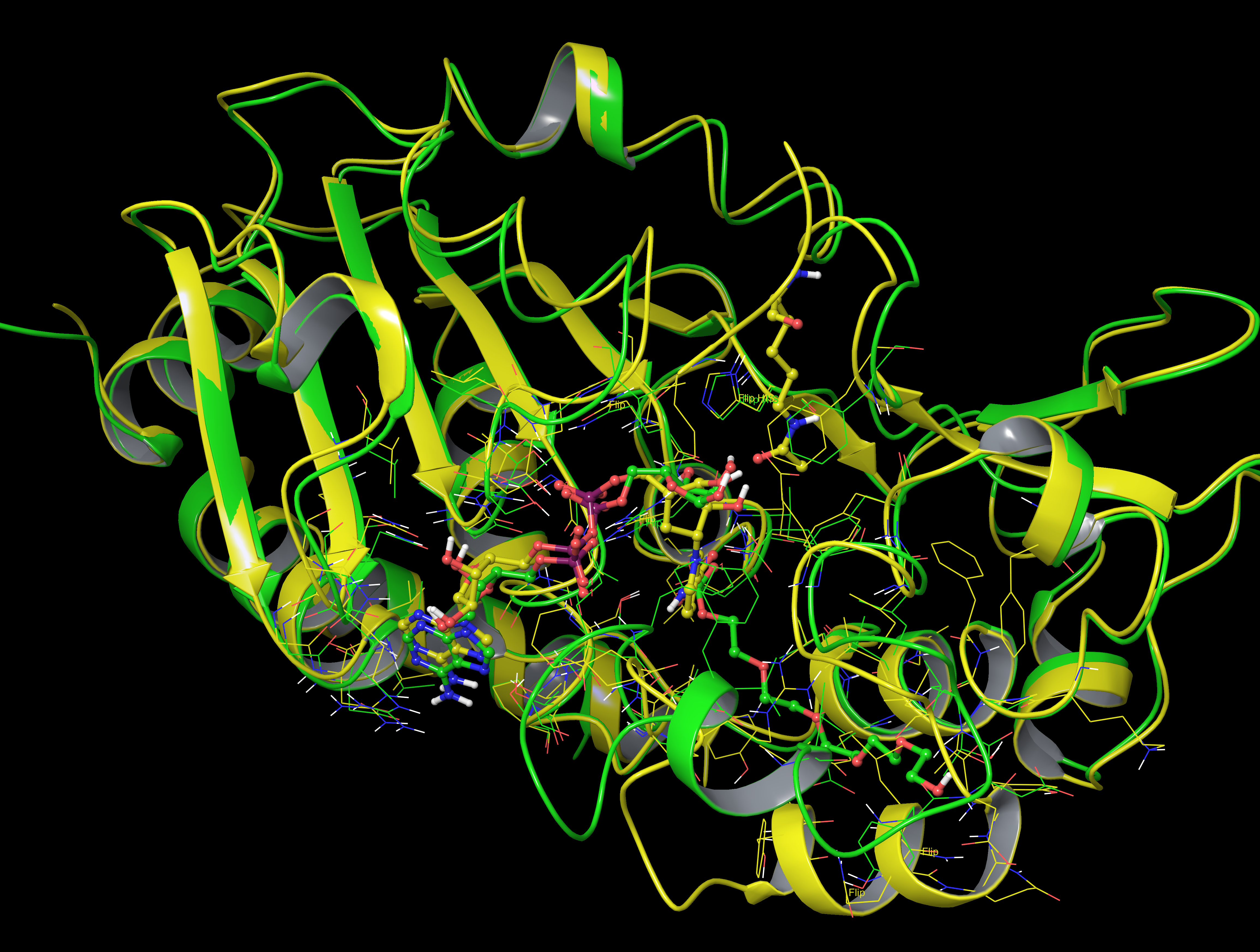
b) Sequence alignment of Sir2Af2 (1YC2), Sir2TM (2H4F), SIRT3 (4FVT), SIRT1 (4I5I).
two chains of 1YC2 were used (chain A and B). and here are the information about the notation in Consensus_aa:
conserved amino acids are in bold and uppercase letters; aliphatic (I, V, L): l; aromatic (Y, H, W, F): @; hydrophobic (W, F, Y, M, L, I, V, A, C, T, H): h; alcohol (S, T): o; polar residues (D, E, H, K, N, Q, R, S, T): p; tiny (A, G, C, S): t; small (A, G, C, S, V, N, D, T, P): s; bulky residues (E, F, I, K, L, M, Q, R, W, Y): b; positively charged (K, R, H): +; negatively charged (D, E): -; charged (D, E, K, R, H): c.
And here is structure alignment between Sir2TM (2H4F, light green, carbon in white) vs SIRT3 (4FVT, yellow, carbon in green ):
and here is the alignment between Sir2TM (2H4F, light green, carbon in white) and SIRT1 (4I5I, with NAD+ and Ex-527, plum, carbon in green):
RC (1-30): I requested the above since we were using Sir2 to represent SIRT1 in the PCB manuscript. Early work referred to SIRT1 as mammalian homolog of Sir2, but there are seven mammalian sirtuin homologs. So we need to be careful about the comparison. Please provide sequence similarity data corresponding to the sequence alignments above for this purpose. Can we do a backbone RMSD comparison for Sir2Tm/SIRT3 and Sir2Tm/SIRT1? Regarding structure alignments, it seems the SIRT1 structure does not have cocrystallized peptide but Sir2Tm (and SIRT3 4FVT?) do. (We may need to use structures without peptide in both cases for proper comparison.)
Have we used Sir2Tm in our manuscript?
RC (1-31): We will probably proceed with NAD and NAM MD binding affinity studies for Sir2Tm next in the presence of peptide (e.g. by docking NAD/NAM to generate starting structures), but I would first like to verify the answers to my latest questions and get the requested info. (In particular, given that most of our existing studies in the paper were done with Sir2Af2, I would like to confirm that there are no Sir2Af2 structures with peptide and that PEG did not induce a similar domain conformational shift as in Sir2Tm).
PL(01/31/14): The above sequence alignment are based on both sequence and structure from crystal structure.
The following sequence alignments with % identity information are based on online program LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html), and structure alignment with backbone RMSD from Schrodinger's suite of program. (numbering based on sequence in crystal structure.)
1) The alignment between Sir2TM (2H4F) and SIRT3 (4FVT):
38.6% identity in 207 aa overlap (19-222:16-212);
RMSD: 2.140 Angstrom
2) The alignment between Sir2TM (2H4F) and SIRT1 (4I5I):
37.3% identity in 212 aa overlap (3-212:3-200);
RMSD: 1.884 Angstrom
3) The alignment between SIRT3 (4FVT) and SIRT1 (4I5I):
42.0% identity in 231 aa overlap (2-227:3-225);
RMSD: 1.888 Angstrom
Details available in the attached file. sequence_alignment_2.docx
Yes. There is no peptide in 4I5I (SIRT1) crystal structure, instead, it has inhibitor (Ex-527) and NAD+. Again, we have very limited crystal structure available for SIRT1, it is hard to say if any Sir2 or other sirtuins can be actually use to structurally represent SIRT1 (and use it to compare with SIRT3).
RC (1-31): Please check the alignment of the A,B,C pocket residues for SIRT1 and Sir2, When we wrote the paper, it was assumed SIRT1 had a B pocket similar to that of Sir2. That can now be verified.
SIRT3 also has a structure with inhibitor (Ex-527) and NAD+ (4BV3) and has a decent alignment with SIRT1 (4I5I).
RMSD: 1.467 Angstrom
No. We did not use Sir2TM structure in the manuscript before.
There is a Sir2Af2 with Ac-p53 (1MA3) and we may be able to combined with 1YC2 (chain B, NAD+ in AC binding mode) to come up with a ternary structure. And B pocket is apparently blocked by the peptide substrate.
RMSD between Sir2Af2 (1MA3) and Sir2Af2 (1YC2:B) is 1.770 Angstrom.
RC (1-31): Ok I will comment on the choice of structure shortly.
RC (2-3): We should use the Sir2Af2 structure above (1MA3) for the Sir2 NAD and NAM MD binding affinity calculations. How do you plan to build the NAD complex - using the superimposed NAD from 1YC2 chain A or docking? For the NAM, through the superposition with chain A for 1YC2 or docking?
Also, regarding the domain motions induced by peptide binding: does 1YC2:B have cocrystallized PEG? (If not, can you provide some RMSD data (or point out data above) that demonstrates this domain motion through an increase in RMSD before/after peptide binding?)
PL(02/04/14): The first step in building complex structure is to complete the Sir2Af2 structure from 1MA3, which has 10 residue missing in the dynamic loop (the 1YC2 may be used as a template in completing the loop). I will try first to build the complex structure by superimpose NAD+ (or NAD+ and NAM both) from 1YC2. They should be good enough as starting structure for MD simulation.
RC (2/04/14): You didn't mention the loop was missing. Are there are residues missing in Sir2Tm? Please post your schedule first as requested (this should be done every week) and I will then comment on Sir2 after getting the updates on other pending tasks. Please also provide the superposition of the pockets (including the loop and NAD.NAM) of 1YC2 and 1MA3.
PL(02/04/14): It is common that some flexible loop can not be resolved in crystal structure, e.g. in Sir2TM ternary structure 2H4F, six residues in the dynamic loop is not resolved. You can easily see the gap in the structures I posted. That's why loop search is a common task in protein structure preparation. Often, the dynamic loop remain flexible in the MD, and stabilized upon ligand binding.
The alignment of 1YC2:B and 1MA3 is attached, including the closer view on the pocket.
RC (2/4/14): I'm aware that flexible loops often cannot be resolved. I have used plop/prime to build flexible loops in the past in work with Friesner. My point was that if we are choosing between Sir2Tm and Sir2Af2 as a starting structure, information on missing residues is relevant to the choice. One is not looking out for all missing residues when looking at the pictures above. The loop building method needs to be discussed. What tools are currently available for this in Prime? We do not necessarily need to use Sir2Af2 because it was used previously, if it is more difficult to build the loop.
PL(02/04/14): Comparing Sir2Af2 (1YC2 and 1MA3) and Sir2TM (2H4F) in term of building ternary structure, the later is certainly much better because it is a ternary structure to begin with, NAD+ and peptide substrate are all in place. 1MA3 doesn't have NAD+ in place, therefore, you can expect extra time for equilibration with NAD+ in place (via either superposition or docking). 1YC2 is lacking peptide and there are requirement for further refinement with peptide substrate in place and both Sir2 and NAD+ might change upon addition of peptide.
The missing loop is 2H4F is not a big issue, only six residues are missing instead of 10 in 1MA3. Schrodinger have tools in Prime to build the loop, and I used to use Modeller for loop modeling.
RC (2/4/13): We may use 2H4F in that case. Prime has ab initio loop prediction capability (sampling to find the minimum energy loop conformation through loop buildup given specified ends of the loop) that is reasonably good for n=6. Does it also allow specification of a template structure? If not, you might try deleting the same loop residues from another pdb file where the complete loop coordinates are known and checking prediction accuracy. With modeller, I believe you would be doing homology modeling with a template structure. Please provide details on the latter method in terms of inputs and outputs.
PL(02/05/14): I will not be using Modeller in this study because its commercial license is issued through Accelrys and is quite expensive to get. I will do the loop search using Prime. And in the MD's early relaxation stage, I will apply a constrain on heavy atoms from crystal structure and let the rest of the system, including the loop, reach equilibration, and refine the loop structure.
RC(02/5/14): Please let me know the RMSD of the Prime-predicted loop to the template structure for both 2H4F and 1MA3. I will then decide on which structure to use.
PL(02/05/14): What do you mean by RMSD of Prime-predicted loop? The loop region is missing, so there is no correct structure to compare to. What is template structure you are referring?
RC (2/5/14): To validate accuracy of loop prediction I initially suggested that you repredict the same loop residues in the structure that you identified as a template above and check RMSD of the predicted to the crystal structure. You mentioned above that 1YC2 may be used as a template in completing the loop. Even if this is not used in the Prime prediction, it could be used for validation of accuracy. You did not reply to the above suggestion, so I suggested above that you consider comparing the Prime predicted loop in 2H4F with the superimposed structure of the loop in 1YC2. The statements above regarding the protocol for loop prediction and MD were also not very clearly written, since you mentioned equilibration and that you would be applying constraints before you indicated you would be doing MD. I assume you will not be applying any constraints in loop prediction (I believe some constraints may be possible in Prime).
PL(02/05/14): For loop search, there will be success and failure. And we might bring up many cases to test Prime's loop search. But I think there is little to do with our approach for modeling. And most often for a loop not resolved in crystal structure due to its flexibility, a single loop structure is not a good representation of the actually structure in solution. I am not sure such validation is really a validation. Yes, I did mention 1YC2 may be used as a template in completing the loop, but that doesn't necessary guarantee a better predicted loop structure, and using MD to refine the loop is probably a better choice. Also, using 1YC2 for validation can be dangerous, because in the nearby flexible region below the C pocket, 1YC2 has a very different loop structure comparing to 2H4F. Using 1YC2 as template for 1MA3 doesn't seem to be a good choice either, although they are both Sir2Af2. The binding of ligand is causing significant change for the flexible region, and they don't align well. My suggestion is, instead of making speculations which loop search method might work better, we use the standard procedure to build the loop and let MD to refine itself.
Also, I want to make clear what tools are available and what each term means in my statement.
Loop search is a homology modeling tool used for building model structure, and currently we use Schrodinger's program and available in Prime (or via protein preparation).
Equilibration, constraints, are terms used for MD simulation. I should probably rephrase the "equilibration" to "relaxation"so you can understand better. There is usually a minimization step and relaxation MD steps in the early stage of the MD simulation before running equilibration and production, in which a constraint is applied to the backbone (or all heavy atoms identified in crystal structure) of the protein and/or heavy atoms of ligands from crystal structure.
I believe constraints could be able to apply in Prime, but it is not obvious from within the Maestro and I don't think it is necessary with out plan in running MD simulations.
RC: Ping, you really don't need to explain the notions of equilibration or homology modeling to me. The issue was with your grammar and that your comments about methodology were insufficiently descriptive. As mentioned, we must agree on methodologies prior to implementing them. You mentioned several approaches and then indicated that you have chosen to apply one without transparency on the method. There was also a choice to be made on what structure to use. I certainly understood your points in retrospect, and yes, I understand that MD simulation can and should be used to refine the loop, but it is not self-evident that a quick validation of the accuracy of the prediction method (which could take as little as a few minutes) is not warranted. Regarding the issue of whether validation is really a validation, there is a whole literature on validation of loop prediction. It is not sufficient to indicate that you built the loop in a particular way later in the manner that you considered best. These points are only specific examples of issues with clarity and transparency on the choice of methodology. We will discuss these points during the performance review, for now please proceed since we do not have time to discuss these points further if they are not already clear.
PL(02/05/14):Sometimes, I don't know what is the information that you are looking for.
So, what is the structure and approach you have decided? Or what other information do you need before making a decision?
RC (02/05/14): Please go ahead and use 2H4F with the approach you indicated, since we do not have time to iterate in this way to compare the accuracy of loop prediction in 2H4F and 1MA3, and due to effects of ligand binding on 1MA3 (the choice of 2H4F or 1MA3 was a decision point, but 2H4F should do).
For future reference, ab initio loop prediction followed by MD is a common protocol. Ab initio loop prediction differs from knowledge-based approaches to loop homology modeling. It uses an energy based sampling method which is faster than MD and hence can be used to rapidly compare structures (like docking) prior to further sampling. With short loops the prediction accuracy can be significant with very low RMSDs to native. It can identify multiple low energy structures if they exist and rank them by MM-GBSA.
Something else you may want to take note of is that the C pocket binding in 1YC2:B (green) is not the same as when peptide are bound. You may see from the figure above that NAD+ configuration is not ready for the first step reaction. And the different is more obvious when compared with Sir2TM (2H4F, Ribbon in yellow, carbon in white ) and SIRT3 (4FVT, cryan).
PL(02/04/14): We may stick with Sir2Af2 because we used it for comparison in the last version of manuscript. However, we need to be careful making connection when we compare the kinetics results of SIRT1 and SIRT3. One significant difference in nicotinamide inhibition between Sir2Af2 and Sir2 (mouse) is that nicotinamide is only partially inhibited by Sir2Af2, whereas in Sir2 mouse, it is complete inhibition.
I will set the Sir2Af2 MD up for running, because not much in-depth MD simulations have been done for sirtuins. (Keun Woo Lee of Korea published a few on human SIRT2). And I think we can also try out simulations for SIRT1.
1YC2:B and 1YC2:C both have PEG in the peptide binding site.(shown below.)
RC (2/4/14): Yes, we are doing Sir2Af2 for a comparison to Sir2, not SIRT1. I am revising the paper to include this comparison. There will now be 3-way comparison. Regarding the differences between SIRT1. SIRT3 and Sir2Af2, I am discussing those in the paper. Hence the structure and sequence analysis tasks for h/mSIRT1 and Sir2Af2 (including questions regarding the C pocket/Met70 in the latter) have been listed on this page. I have also asked a question above regarding domain motions induced by peptide binding, which is relevant to hSIRT1 structure.
PL(02/04/14): I assume the domain motions by peptide binding refer to the change in Sir2Af2 vs with PEG binding.
It appears that with peptide bound, the two domains in Sir2Af2 does get closer to each other. In 1YC2 chain B and C (NAD+ in C pocekt) has a better RMSD (1.77 Angstrom) vs 1MA3 compared to 1YC2 chain A and D (2.02 Angstrom). And structural alignment is shown below. (1YC2:B in green, 1MA3 in purple)
RC (2/4/14): Can you provide an RMSD for A,B,C pocket residues only, since those are the ones most relevant to catalysis?
RC (2/6/14): And, how about a similar analysis for SIRT3 with and without peptide, if the appropriate structures are available?
PL(02/07/14): Several attempts to align only the residues of A,B,C pocket all failed in Schrodinger. I may try again some other time using other software.
The comparison for crystal structures of SIRT3 w/ or w/o peptide substrate is performed using 4FVT and 4BN4 (SIRT3 with ADP-Ribose). The later is the best we can found to represented bound NAD analog without the presence of peptide substrate. There is a PEG with and ending acetic acid bound in the C pocket in 4BN4. The RMSD between 4FVT and 4BN4 is 1.473 Angstrom, and the residues in the A binding pocket from the Rossmann-fold domain align up very well between the two.
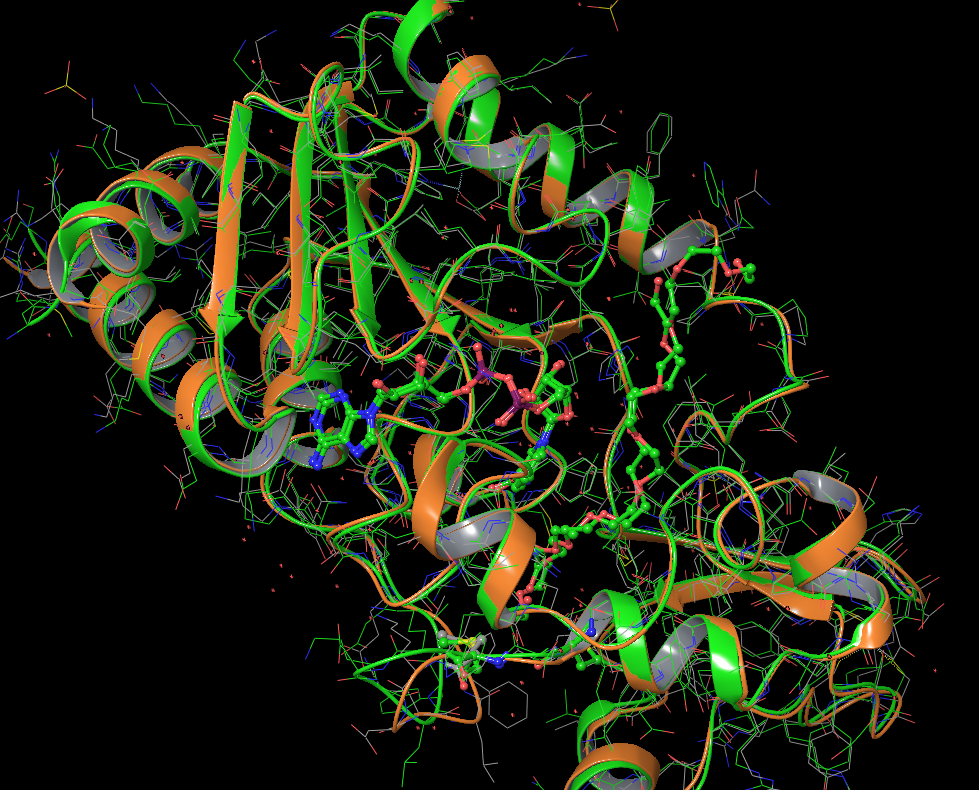
For comparison, RMSD between chain A and chain B in 1YC2 is 1.805 Angstrom. Chain A has NAM and NAD+ in AB pose.
c) In Sir2Af, when NAD+ bound to AC pocket as in 1YC2 (chain B and C), Met70 is not in the C pocket. For comparison, aligned structure between Sir2TM (2H4F. yellow, Met71 in ball and stick, carbon in white) and Sir2Af (1YC2 chain B, green, Met70 and a nearby Met68 in ball and stick, carbon in green.)
PL(02/04/14): A few more words on Met70: In Sir2TM (2H4F) Met71 is in C pocket. However, Met70/Met68 are not in the C pocket in 1YC2, and it is getting closer to the C pocket (not in though) in the structure of 1MA3. From the structure alignment we do not find Met in the same position as in Sir2TM in SIRT3 or SIRT1. This feature may be unique to Sir2TM (or some other Sir2).
Situation is similar for 1YC2 chain A, in which NAD+ in AB pocket, Met70 is not in C pocket.
RC (1-30): I requested the above to determine if Met70 may be affecting NAM binding in Sir2Af2. In chain A, is Met70 close to the NAM? Have you already done a docking study of NAM to this Sir2Af2 C pocket? If so, please show the result.
RC (1-31): It appears there is a cocrystallized structure of Sir2Af2 with NAM in the C pocket, so analysis of that structure (instead of docking) should be sufficient.
I believe the Sir2Af2 structures above do not have cocrystallized peptide (I believe there is only one Sir2Af2 structure with a PEG molecule in the peptide binding pocket). As I understand, there is no analogous Met in Sir2Tm.
RC (1-31): Sir2Af2 has less sensitivity to nicotinamide inhibition than other sirtuins; Sauve claims this is due to the base exchange equilibrium constant not the Kd for NAM. It will be interesting to study the effects of side chain variations in/near the C pocket on the conformation of NAM. This is not yet a priority for MD simulations, but please comment on whether the NAM MD trajectories obtained for SIRT3 C pocket approach the region wherein Met70/Met68 are located in Sir2Af2 (since those are the only MD trajectories we have for NAM at this time).
PL(01/28/14): schedule for week 01/27/14-01/31/14:
1) continue MD and MM-PB(GB)SA calculations for interested systems (currently MD for SIRT3/AcCS2/NAD+; SIRT3/AcCS2/NAD+/NAM; SIRT3/AcCS2/NAD+/isoNAM are completed. SIRT3/NAD+ (NAD+ start from AB and AC pose) are still running, SIRT3/AcCS2/NAD+/1-methyl-NAM is scheduled. Short MD simulations for SIRT3, AcCS2, and NAD+ alone are completed. MM-PB(GB)SA calculations using three trajectories will be scheduled.
RC (1/28): Ok. You need to indicate in advance the simulations that are being carried out, if they have not been discussed and especially if they are related to the methodology. You previously indicated that the free energies of the ligand+apoprotein for Delta G calculations are being based on the same trajectories as the complex but by turning off the protein-ligand interactions. Are the short MD simulations to be used for free ligand free energy calculations? By three trajectories, do you mean those for the free ligands?
Please let me know once the SIRT3/NAD+ MD's are completed, and roughly how long the MM-PB(GB)SA analysis will take. We may prefer to run some Sir2 simulations prior to 1-methyl-NAM. For that, the task above on the Sir2 sequence/structure alignments (and task c above) is necessary. If you have any questions on which structures to use, let me know. I will provide more information on possible MD simulations after receiving the above.
PL(01/29/14): Yes. the short MD simulations are to be used for free energy calculation using free ligand instead of the bound form. So far, all the analysis reported (Delta G calculations) are based on single trajectory MD simulations, as it is a common practice in MM-PB(GB)SA calculations. However, simulations using three trajectories, namely, MD simulations for complex, receptor, ligand separately, will produce more accurate results. You addressed the concern before, and that's why I carried out the simulation and if the single trajectory approach is not working correctly, and we can use the results from new analysis using three trajectories to find the clues.
As you can see from SIRT3/NAD+ simulation, the interactions shows large fluctuations. And by checking the trajectory, I found apparent deviation from the starting structure. I expected it may take longer to reach equilibration or the fluctuation is an intrinsic property of the system, I will keep you updated on the progress.
I haven't start simulation for 1-methylnicotiname yet, and will check out Sir2 first.
PL(01/30/14): Some updates from SIRT3/NAD+ MD simulation:
NAD+ doesn't bound tightly to SIRT3, as we can see from MD simulation.
Starting from NAD+ in AC pocket (initial structure build from ternary structure of 4FVT), snapshots taken at 0ns, 10ns, 20ns, 22ns, 24ns, 26ns, and we found that while adenine nucleotide moiety still binds to the A pocket, the nicotinamide moiety fluctuate a lot. After 20 ns, the nicotinamide moiety is completed out of C pocket.
NAD+ doesn't bind strongly to the B pocket either. Starting from NAD+ in AB pocket (initial structure build by superimpose NAD+ from 1YC2 (chain A) to SIRT3 taken from 4FVT), snapshots were taken at 0ns, 4ns, 8ns and 12ns, and we found that the nicotinamide moiety continue to swing from enzyme and mostly expose itself to water solvents.
SIRT3_NADp_MD_snapshots.docx
RC (1-31): Do we need to run a longer MD simulation to get an "accurate" binding affinity estimate (i.e. one that is sampling from equilibrium distribution and insensitive to initial conditions) for NAD:SIRT3 in AC pocket? Please provide the binding affinity convergence data when you have it. How does NAM moiety trajectory in SIRT3:NAD MD simulation compare to that for SIRT3:NAM?
Raj, what Sir2 systems are you interested of running MD? I can start preparing the structures.
2) carry out structural analysis on the MD results, i.e. RMSD and B-factor calculations, comparing averaged MD structure with crystal structures.
RC (1/28): Regarding RMSD, are you referring to comparison between the MD averaged ligand structure and the co-crystallized ligand structure? If so, for which complexes is this possible? Please let me know if you are planning to do this for the peptide and NAD complexes for which structures are available in order to benchmark the MD sampling method.
Are you going to prepare figures based on the averaged MD structures that can be used in the paper, to replace the docking figures? Among these, please prioritize the NAM/isoNAM structural analysis, followed by that for NAD, with peptide being lowest priority. Please send me the complete NAM/isoNAM results when finished so I can consider whether any changes are required prior to using them in the paper.
PL(01/29/14): The RMSD I mentioned refer to the backbone of SIRT3, which shows structural changes from the starting structure (e.g. 4FVT). I have no plan in calculating RMSD of ligands.
As to what figure to report, it highly depends on what information is useful in supporting our results. I will be analyzing structures from MD simulations and will report them if there is something useful in supporting Delta G results.
RC (1-31): I will be inquiring further about possible figure replacements shortly. Since we will likely be replacing at least several of the existing figures generated by docking, I would like to see some of the averaged MD structures of the complexes for which we have been calculating binding affinities.
3) force field parameters and topology are ready for intermediate, MD can be scheduled if needed.
4) draft for computational methods
5) work on the tasks provided.
RC (1/28): Some of the ongoing work involves long simulations during which there may be time for other work. Please indicate how much time there is in the schedule over the next 1-2 weeks (if any) for PCR tasks.
PL(01/29/14): Result/data analysis is intensive and time-consuming, and I also have to monitor the MD simulations (check out trajectory structures and run MM-(PB)GBSA calculations) to decide if I should extend the MD. I have only very limited time for the PCR tasks so far, but once I get time, I will work on it. By the way, did you find any useful files from your past collections?
PL(01/14/14):
1) SIRT3/peptide/NAD/isoNAM is currently running. I will soon provide an update if everything is all right.
2) Attached please find the reference requested. Mechanism of Nicotinamide inhibiton and transglycosidation by Sir2_2003_Jackson.pdf
RC (1-15): Though there isn't a great correlation between pyridine ring nitrogen pKa and base exchange, please look into comparison of isoNAM and NAM pyridine N pKa using either Epik (empirical) or qm methods. In this regard please provide the qm charge fits for NAM and isoNAM. Also, please start running the qm charge fitting for the tightest binding C pocket inhibitor from XG's experiments on a cpu node.
PL (01/21/14): QM Charges, and atom types for NAM and isoNAM in the Amber prep file format (including topology)
NAM's prep file
isoNAM's prep file:
PL(01/23/14): Charges, atom types and topology of 1-methylnicotinamide in Amber prep file format:
RC (1-21): I assume the methods for pKa estimation above are either too inaccurate (Epik) or too time consuming to warrant pursuing at this time? Please let us know which N is the one of interest (pyridine ring N) above.
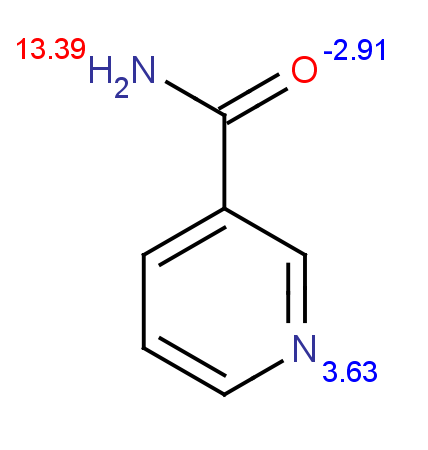
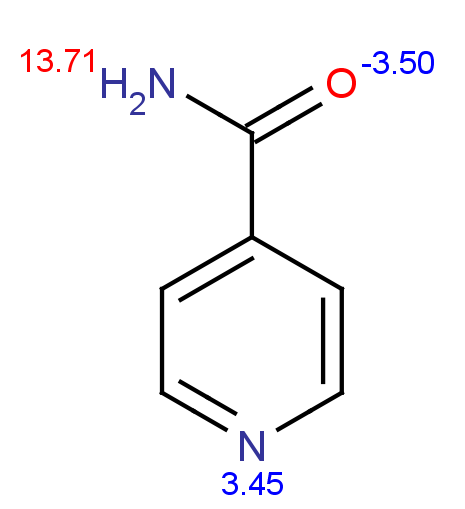
PL(01/21/14): I haven't had time to look into the pKa calculation/estimation for NAM and isoNAM. The N1N above is the one of interest.
PL(01/21/14):Here are the predicted pKa for NAM (left) and isoNAM (right) using the following web service:
http://epoch.uky.edu/ace/public/pKa.jsp (they use chemaxon's JChem Suite for empirical prediction.)
3) It is possible to run MD simulation for the binding of NAD in AC pocket of SIRT3.
RC (1-15): Ok, in that case this simulation may take priority over the Sir2 simulations above. Please comment on the differences in the A pocket and NAD contacts (identified e.g. by docking) induced by peptide binding to SIRT3. Please make preparations to run this simulation while waiting for the isoNAM simulation to run.
PL(01/21/14): In order to compare the different, one possible way is to compare SIRT3/Peptide/NAD+ with SIRT3/NAD+, the later can only be obtained through MD. The other possible way is compare SIRT3 vs. SIRT3/Acetylated Peptide, (3GLS vs 3GLR), and see the structural changes, then carry out docking of NAD+. The later approach is not accurate since we observed further structural changes upon binding of NAD+ (4FVT), especially to the open end of A pocket.
And here is the comparison between apo-enzyme (3GLS, red) and SIRT3 with Ac-Peptide (3GLR, yellow): Upon binding of peptide, the two domain do get closer.
RC (2-4): Please provide relevant RMSDs regarding the domain motion.
Here is the same figure with backbone only illustration:
However, upon NAD+ binding, (4FVT, purple), the flexible loop changes its conformation and getting more contact with A pocket.
RC (1-21): Ok, in that case we can look at the MD statistics during the equilibration and compare them to the previously acquired statistics in the presence of peptide, in order to get an idea of the flexibility
of the NAD binding mode in the absence of peptide. I would also be interested in seeing how the loop moves during MD. Please provide these statistics when they are available.
Based on the info below, since you are equilibrating for only 1.8ns, this comparison can be provided during the initial several ns of the simulation.
4) See 3).
5) After checking that there is nothing wrong with the setup and parameterization in the SIRT3/peptide/NAD/isoNAM simulation, I will let it run for one week. After that, MM-PB(GB)SA calculations will be carried out to estimate the binding affinity.
RC (1-15): Please provide me with convergence data on this simulation as it is available. I would like to get an idea of how long it takes for equilibration and the rate of convergence of the binding affinity estimate, so we can determine the appropriate duration of MD simulations given our available timeframe.
PL (01/21/14): MM-PB(GB)SA calculation is very time-consuming because it use only one processor at a time. MM-PBSA values are usually closer to experimental values, but MM-GBSA calculations are much faster.
Here are calculated MM-PB(GB)SA values for every 2 ns (first 1.8 ns not used, frames taken every 10 ps, no entropy correction, error bar from statistics).
RC (1-21): Regarding one processor, I assume this is due to the Schrodinger license. How long did the MM-PBSA calculations below take? Higher throughput MM-PB(GB)SA calculation was one application I had in mind for scripting with free software tools. Would this allow us to use an arbitrary number of processors?
PL (01/21/14): No, I am not using Schrodinger's module in calculating MM-PB(GB)SA here. I am using the free AmberTools instead, There are 200 frames in 2 ns trajectory ( 10 ps sampling step), and GBSA and PBSA combined takes about eight hours to complete.
PL (01/23/14): To be more specific, MM-GBSA for 200 frames takes about 1 hour, and MM-PBSA for 200 frames takes about 7 hours. I have been looking into the tools or developing a tools to use AMBER to score and rank docking poses. DOCK seems to have tools for doing that, but it costs quite a bit for commercial use.
RC (1-21): Ok; in that case why can only one processor be used at a time to compute MM-PB(GB)SA energies? It that because AmberTools automates the calculation and it is tedious to split the computation between nodes?
PL (01/23/14): MMPBSA.py code in AmberTools supports MPI, but since I haven't installed MPI on the GPU cluster, I didn't compile the MPI version for parallel calculations. However, I am breaking down into multiple sections in the calculations which can run simultaneously, and the overall mean and standard deviation can be derived by merging the results from sections.
RC(1-21): Please provide some more info on the data below. a) are the error bars obtained from the sample set of 10 ps frames in the window between the specified time in ns and the previously specified time in ns?
b) Is this post-equilibration? If so how long did equilibration take? Did you monitor the B factor during equilibration and if so how did you assess convergence to equilibrium so as to set equilibration time?
c) How long did it take to obtain the 25 ns trajectory?
d) For convergence analysis of the binding affinity, I was interested in the convergence of the MM-PB(GB)SA ensemble estimate for the binding affinity based on all the data up to a particular time. Is this being prepared?
When you get a chance, please also post the analogous data for the NAM system.
PL(01/21/14): a) error bars are from 200 frames in every 2 ns simulation.
b) I do not have time to carry out analysis to confirm equilibration, so I simply set up MM-PB(GB)SA calculations from 1.8 ns.
c) the 25 ns simulation was done last week (whole week).
d) there will be too time consuming to calculate both GBSA and PBSA using 10 ps sampling step, therefore, I am about to prepare to get convergence on GBSA calculation only.
Convergences calculation for MM-PB(GB)SA calculations for isoNAM is planned.
PL(01/23/14): More convergence information is followed. Again, first for the NAD+ binding as seem above. Only MM-GBSA results (from 1.8 ns up to t_end) are reported here.
RC (1-23): If I interpret this convergence data properly, it appears that your estimate is not changing significantly after 10-15 ns. Certainly 20 ns is sufficient. Will be interesting
see the convergence results for the other complexes. I was primarily interested in using these convergence analyses to justify the choice of simulation time in our binding affinity calculations.
The correlation between MM-GBSA and MM-PBSA is not very good.
The above MM-GBSA and MM-PBSA results can easily be obtained from the 2ns analysis.
From the same trajectory, the results is better if we run statistics after 5.8ns. (As it is obvious from the data above.)
I also did a analysis using 20 ps sampling step from 5.8 ns up to t_end. And the MM-GBSA results is here:
I also did the same analysis for Peptide binding and isoNAM binding, here only the results:
RC (1-23): Is t_start above the same as t in the earlier raw data for NAD? If I understand correctly you have not yet run convergence calculations for peptide and isoNAM.
(BTW, for peptide, there is an issue in terms of computation of binding affinity, due to change of protein conformation on peptide binding.)
PL (01/23/14): Yes. t_start here is the same as t earlier. I think it is a more appropriate description.
I can generate the same plot for up to certain t_end from the 2 ns analysis. I presented it this way because I think it tells more about the fluctuations of structure and binding, and also be used to help locate the approximate point of equilibration. (Currently, I did not specify the time used for equilibration because it depends significantly on the starting structure of the system.)
The binding affinity calculation using the single trajectory does have limitation in terms of accuracy, as we didn't include the free energy cost for conformation change, and the entropy contribution, and the impact of force field and GB or PB model. We can probably get an estimate based on previous studies on other systems from literature.
PL (01/23/14): Here are the convergence plot you expected for peptide and isoNAM binding. As you can see many details about the fluctuations got lost as they always converge to the averaged value.
6) MM-PB(GB)SA protocol will be provided soon.
PL(01/21/14): Schedule for week 01/20/14-01/24/14:
1) continue MD simulation with SIRT3/peptide/NAD/isoNAM and carry out MM-PB(GB)SA calculation.
2) the QM calculation completed for 1-methylnicotinamide cation, charge fitting and force field preparation will be carried out.
3) MD simulations for SIRT3/NAD binary system is currently running, and MM-PB(GB)SA will be carried out alone the way to test the convergence.
4) Setting up MD simulation with Sir2Af2/NAD (1YC2) in case we do need to run the simulation
5) draft the method section to include the charge fitting procedure and results, and MM-PB(GB)SA calculation procedures.
6) structural analysis on MD simulations
RC (1/21): The draft methods section and preliminary figures will be important, since I would like to consider how they will fit into the structure of the revised paper while we keep generating results.
RC (1/20): PL and XG, please provide (separately) your schedules for this week vis-a-vis the pending tasks above. PL, please include a schedule for PCR software work. (PL: PCR software survey will be updated on Ping's task list).
RC (1/21): Please provide answers to the questions from last week in bold above. In order to properly manage the project, it is important that I am able to see results as they are generated so that if necessary, modifications to the plan can be made on the fly based on the results. Examples include a) the convergence statistics for the MD simulations; without this info, it is not clear how long these simulations should be run and why we have chosen to run them for a particular time. b) Summary of the structural differences between the apo- and peptide-bound A pocket structures of SIRT3, which will determine whether binding affinity calculation on the former will be useful to include in the paper - e.g., does the A pocket close on peptide binding? In this regard the early MD statistics during the equilibration of this complex may be useful to compare to those from the SIRT3/peptide/NAD complex.
This is especially important given the time taken to run MD simulations. In general, if the requested information is not available please indicate when it will be available or why it is not available.